Two-point flux-approximation (TPFA) for Darcy flow
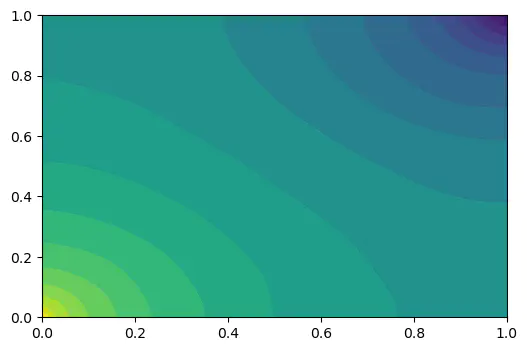 Results from this method
Results from this methodProjects description
Purpose: This is my notes for using Matplotlib to make animation, it’s very convenient during your simulation.
1.1 Functions
1.1.1 Basic Functions
Continuity function
Darcy Law
1.1.2 Instance: for water
Simplicity:
Calculating the divergence for
From eq. (1.1):
and from eq. (1.2):
So, we can get equation (1.3), where the lower label
Bundary condition on the reservoir boundary
Understanding Let’s take a look at Eq. (1.3), on the left side (the first equality),
1.2 TPFA(Two-point flux-approximation)
TPFA scheme, is a cell-centred finite-volume method, which is also one of the simplest discretization techniques for elliptic equations.
1.2.1 Functions of the
Obviously, it’s coming from the left two term of Eq.(1.5).
- Assuming that
Attention:
is the volume of one finite volume(or a cell), stands for the surface of one finite volume(or a cell), and is the interfaces of two neighbor cells.
- Summary
Obviously, the left term in Eq.(1.6) is unknown (the
1.2.2 Functions the
Making
Reformulate Eq.(1.3) to get the proper equation like the Eq.(1,6), which is the standard TPFA FVM scheme.
Integrate over the finite volume
Apply the divergence theorem
As ,the u_w only changes between cells, the
So, finally, we can get:
$$\int {\Omega}\frac{q_w}{\rho_w}d\Omega=\int {\gamma{ij}}(\lambda \bigtriangledown u_w)d\gamma{ij} \tag{1.11}$$
1.2.3 Discretization
Let’s discretization with Eq. (1.11) and adding a flux term
Discretization of
Discretization of
Discretization of Eq. (1.11):
$$v_{ij}=-|\gamma {ij}|\lambda{ij}\delta u_{ij}=2|\gamma _{ij}|(\frac{\bigtriangleup x_i}{\lambda _{i,ij}}+\frac{\bigtriangleup x_j}{\lambda _{j,ij}})^{-1}(u_i-u_j) \tag{1.14}$$
1.2.4 Solve the
Simplize Eq.(1.14) Terms that do not involve the cell potentials
Solve the function According to Eq. (1.15), only
1.2.5 Solve the
Function According to Eq.(1.2) and with some new parameters we defined during the process.
Discretization According to Eq. (1.12), we can get:
Solve the function As the
1.3 Code
1.3.1 Numpy version
# -*- coding: utf-8 -*-
"""
Created on Sun Aug 7 11:00:33 2022
@author: Howw
"""
import numpy as np
import matplotlib.pyplot as plt
import scipy.ndimage
from scipy.sparse import spdiags
from struct_tools import DotDict
np.random.seed(42)
# TPFA(two-point flux-approrimation) finite-volume discretisation
def TPFA(Nx,Ny, K, q):
# Compute transmissibilities
hx,hy=1/Nx,1/Ny
L = K**(-1)
''' this is the t_ij in function(see notes),
as cell in current problem is same-wise (in the same direction, so \delta xi= \delta xj)
and for 2-D problem, the area of cell equals to dx or dy.
'''
TX = np.zeros((Nx+1, Ny))
TY = np.zeros((Nx, Ny+1))
TX[1:-1, :] = 2*hy/hx/(L[0, :-1, :] + L[0, 1:, :])
TY[:, 1:-1] = 2*hx/hy/(L[1, :, :-1] + L[1, :, 1:])
# Assemble TPFA discretization matrix, Ravel is better than reshape(N,1)
x1 = TX[:-1, :].ravel()
x2 = TX[1:, :] .ravel()
y1 = TY[:, :-1].ravel()
y2 = TY[:, 1:] .ravel()
## Setup linear system(confused?)
DiagVecs = [-x2, -y2, y1+y2+x1+x2, -y1, -x1]
DiagIndx = [-Ny, -1, 0, 1, Ny]
## Coerce system to be SPD
DiagVecs[2][0] += np.sum(K[:, 0, 0])
A = spdiags(DiagVecs, DiagIndx,N,N).toarray()
## Solve to get all u
u = np.linalg.solve(A, q)
# Get fluxes, V, of each cell
P = u.reshape(Nx,Ny)
# use DotDict from sturct_tool to make V become 2-D with matrix
V = DotDict(
x = np.zeros((Nx+1, Ny)),
y = np.zeros((Nx, Ny+1)),
)
V.x[1:-1, :] = (P[:-1, :] - P[1:, :]) * TX[1:-1, :]
V.y[:, 1:-1] = (P[:, :-1] - P[:, 1:]) * TY[:, 1:-1]
# Get velocity of each cell
v=DotDict(
x = np.zeros((Nx+1, Ny)),
y = np.zeros((Nx, Ny+1)),
)
v.x[1:-1,:]=-TX[1:-1,:]*(P[1:,:]-P[:-1,:])/(hx)
v.y[:,1:-1]=-TY[:,1:-1]*(P[:,1:]-P[:,:-1])/(hy)
return P, V, v
Nx,Ny=32,32
N=Nx*Ny
# K=np.ones((3,8,8))
# K=np.exp(5* ((np.random.rand(3, Nx, Ny))))
K=np.exp(5* scipy.ndimage.uniform_filter((np.random.rand(3, Nx, Ny)),5))
q=np.zeros(N)
q[0]=1
q[-1]=-1
#p is the pressure, V is the flux, v is the velocity
P,V, v=TPFA(Nx,Ny,K,q)
#draw the results
levels=np.linspace(P.min(),P.max(),20)
Grid_X=np.linspace(0,1,Nx)
Grid_Y=np.linspace(0,1,Ny)
plt.contourf(Grid_X,Grid_Y,P,levels=levels)
plt.savefig("./figures/p.png",dpi=100,bbox_inches="tight")
plt.contourf(Grid_X,Grid_Y,v.x[:-1,:])
plt.savefig("./figures/vx.png",dpi=100,bbox_inches="tight")
plt.contourf(Grid_X,Grid_Y,v.y[:,:-1])
plt.savefig("./figures/vy.png",dpi=100,bbox_inches="tight")
1.3.2 Torch version
# -*- coding: utf-8 -*-
"""
Created on Sun Aug 7 11:00:33 2022
@author: Howw
"""
import numpy as np
import torch
import matplotlib.pyplot as plt
import scipy.ndimage
from struct_tools import DotDict
import time
random_seed=42
#this function is based on torch, acted like scipy.sparse.sodiags
#data should be 1-D,and diags should be an int
def torch_spdiags(data,diags):
data_out=torch.zeros(data.shape[0],data.shape[0])
if diags==0:
data_out=data_out+torch.diagflat(data,0)
if diags>0:
data_out=data_out+torch.diagflat(data[diags:],diags)
if diags<0:
data_out=data_out+torch.diagflat(data[:diags],diags)
return data_out
# TPFA(two-point flux-approrimation) finite-volume discretisation
def TPFA(Nx,Ny, K, q):
# Compute transmissibilities by harmonic averaging.
hx,hy=1/Nx,1/Ny
L = K**(-1)
TX = torch.zeros((Nx+1, Ny))
TY = torch.zeros((Nx, Ny+1))
TX[1:-1, :] = 2*hy/hx/(L[0, :-1, :] + L[0, 1:, :])
TY[:, 1:-1] = 2*hx/hy/(L[1, :, :-1] + L[1, :, 1:])
# Assemble TPFA discretization matrix, Ravel is better than reshape(N,1)
x1 = TX[:-1, :].ravel()
x2 = TX[1:, :] .ravel()
y1 = TY[:, :-1].ravel()
y2 = TY[:, 1:] .ravel()
## Setup linear system(confused?)
## Coerce system to be SPD (ref article, page 13).
## Version 1: failed in always using torch
##Attention: as I failed in find spdiags in Torch, Here the SPD is still using scipy and numpy
# DiagVecs = [-x2.numpy(), -y2.numpy(), (y1+y2+x1+x2).numpy(), -y1.numpy(), -x1.numpy()]
# DiagIndx = [-Ny, -1, 0, 1, Ny]
# A_temp=spdiags(DiagVecs, DiagIndx,N,N).toarray()
# A = torch.from_numpy(A_temp)
## Version 2:successfully using torch all the time by self-defined function
DiagVecs =[-x2, -y2, (y1+y2+x1+x2), -y1, -x1]
DiagVecs[2][0] += torch.sum(K[:, 0, 0])
A=torch_spdiags(DiagVecs[0],-Ny)+torch_spdiags(DiagVecs[1],-1)+\
torch_spdiags(DiagVecs[2],0)+torch_spdiags(DiagVecs[3],1)+torch_spdiags(DiagVecs[4],Ny)
## Solve
u = torch.linalg.solve(A, q)
# Extract fluxes
P = u.reshape(Nx,Ny)
# use DotDict from sturct_tool to make V become 2-D with matrix
V = DotDict(
x = torch.zeros((Nx+1, Ny)),
y = torch.zeros((Nx, Ny+1)), # noqa
)
V.x[1:-1, :] = (P[:-1, :] - P[1:, :]) * TX[1:-1, :]
V.y[:, 1:-1] = (P[:, :-1] - P[:, 1:]) * TY[:, 1:-1]
# Get velocity of each cell
v=DotDict(
x = torch.zeros((Nx+1, Ny)),
y = torch.zeros((Nx, Ny+1)),
)
v.x[1:-1,:]=-TX[1:-1,:]*(P[1:,:]-P[:-1,:])/(hx)
v.y[:,1:-1]=-TY[:,1:-1]*(P[:,1:]-P[:,:-1])/(hy)
return P, V,v
#loss function based on Eq. 1.17
def loss_TPFA(P,v,Nx,Ny,K):
hx,hy=1/Nx,1/Ny
L = K**(-1)
Lamada_x = torch.zeros((Nx+1, Ny))
Lamada_y = torch.zeros((Nx, Ny+1))
Lamada_x[1:-1, :] = 2*hy/hx/(L[0, :-1, :] + L[0, 1:, :])
Lamada_y[:, 1:-1] = 2*hx/hy/(L[1, :, :-1] + L[1, :, 1:])
loss_x=v.x[1:-1,:]+Lamada_x[1:-1,:]*(P[1:,:]-P[:-1,:])/hx
loss_y=v.y[:,1:-1]+Lamada_y[:,1:-1]*(P[:,1:]-P[:,:-1])/hy
return loss_x,loss_y
#Initializing
Nx,Ny=64,64
N=Nx*Ny
# K=np.ones((3,8,8))
# K=np.exp(5* ((np.random.rand(3, Nx, Ny))))
K=np.exp(5* scipy.ndimage.uniform_filter((np.random.rand(3, Nx, Ny)),5))
K=torch.from_numpy(K)
q=torch.zeros(N)
q[0]=1
q[-1]=-1
start = time.perf_counter()
P,V,v=TPFA(Nx,Ny,K,q)
end = time.perf_counter()
print("Time:",end-start)
loss_x,loss_y=loss_TPFA(P,v,Nx,Ny,K)
#Draw the distribution of pressure
levels=np.linspace(P.min(),P.max(),20)
Grid_X=np.linspace(0,1,Nx)
Grid_Y=np.linspace(0,1,Ny)
# plt.contourf(Grid_X,Grid_Y,P,levels=levels)
# plt.contourf(Grid_X,Grid_Y,P,levels=levels)
# plt.savefig("./figures/p.png",dpi=100,bbox_inches="tight")
# plt.contourf(Grid_X,Grid_Y,v.x[:-1,:])
# plt.savefig("./figures/vx.png",dpi=100,bbox_inches="tight")
# plt.contourf(Grid_X,Grid_Y,v.y[:,:-1])
# plt.savefig("./figures/vy.png",dpi=100,bbox_inches="tight")
1.4 Loss function
1.4.1 Description
As this work is used for generating data for Operator training, it’s better to difine a loss function in advance, which can help the training process as combined with MSE.
1.4.2 Definition
In this work, only one function ban be uesd as loss function as there is no useful initial condition of
Let’s take a look at code for calculating
v=DotDict(
x = np.zeros((Nx+1, Ny)),
y = np.zeros((Nx, Ny+1)),
)
v.x[1:-1,:]=-TX[1:-1,:]*(P[1:,:]-P[:-1,:])/(hx)
v.y[:,1:-1]=-TY[:,1:-1]*(P[:,1:]-P[:,:-1])/(hy)
And showing the process with sketching:

So, only function (1.17) can be uesd as loss function, as other functions, like Eq. 1.1, containing q(Nx*Ny), which can’t be used with
1.4.3 Code
#loss function based on Eq. 1.17
def loss_TPFA(P,v,Nx,Ny,K):
hx,hy=1/Nx,1/Ny
L = K**(-1)
Lamada_x = np.zeros((Nx+1, Ny))
Lamada_y = np.zeros((Nx, Ny+1))
Lamada_x[1:-1, :] = 2*hy/hx/(L[0, :-1, :] + L[0, 1:, :])
Lamada_y[:, 1:-1] = 2*hx/hy/(L[1, :, :-1] + L[1, :, 1:])
loss_x=v.x[1:-1,:]+Lamada_x[1:-1,:]*(P[1:,:]-P[:-1,:])/hx
loss_y=v.y[:,1:-1]+Lamada_y[:,1:-1]*(P[:,1:]-P[:,:-1])/hy
return loss_x,loss_y
1.5 Results
1.5.1 The distribution of pressure

1.5.1 The distribution of velocity


Reference
- G. Hasle, K.-A. Lie, E. Quak, and Selskapet for industriell og teknisk forskning ved Norges tekniske høgskole, Eds., Geometric modelling, numerical simulation, and optimization: applied mathematics at SINTEF. Berlin ; New York : [Oslo]: Springer ; SINTEF, 2007.